Jungle Rhythms workflow
Koen Hufkens
2023-12-19
workflow.RmdPre-processing
The Jungle Rhythms workflow relies on manual template. In particular, it addresses the issue of outlining locations in a table which contain data, and reducing the complexity of the transcription.
Manual template outlines
As such the workflow will generate data which only presents one unit of interest (a year) at a time for annotation, limiting the chance of propagating errors in incomplete or corrupted series. At the same time this makes the task easier to complete in informal settings, on a cellphone or a tablet rather than a computer.

A single year, cut out for processing
Zooniverse project setup
The Jungle Rhythms project was run on the Zooniverse platform. The platform provides an easy project builder which allows you to specify how annotations should be made on images. An in depth description of our setup is outside the scope of this workflow as project requirements will vary greatly. We refer to Zooniverse project builder documentation and their excellent forums to help you setup a project.
Post-processing
Data was collected from the Zooniverse project builder interface as
complex json file (but stored in the R specific rds format
to save space). For each section shown to multiple citizen scientists we
distilled the coordinates (i.e. timing) of phenology events through a
number of basic geometry operations (line intersections, coordinate
transformations etc). Functions in support of this post-processing are
provided by this package and stored in the R folder. The processing
workflow of this raw data is stored in the data-raw folder
(published online in due course).
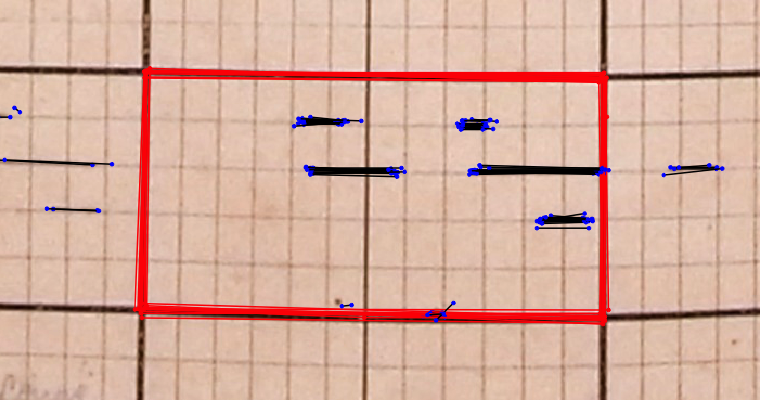
Individual annotations as made by citizen scientists, to be processed in consistent phenoloogy observations.

